Eukaryotic genomes such as yeast were used to train EVO-2 models.Credit: Thomas Deerinck, NCMIR/Science Photo Library
Scientists today announced that what they say is the largest artificial intelligence (AI) model ever for biology.
The model was trained with 128,000 genomes ranging from humans to single-celled bacteria and archaea, allowing the entire chromosome and small genomes to be written from scratch. It also allows us to understand existing DNA, including “non-encoding” gene mutants associated with the disease.
“CHATGPT for CRISPR” creates a new gene editing tool
The EVO-2, co-developed by researchers from ARC Institute and Stanford University, from Palo Alto, California and chip maker Nvidia, is available to scientists via a web interface. I had to replicate the model.
Developers see EVO-2 as a platform that others can adapt to their uses. “We are really looking forward to how scientists and engineers can build this ‘app store’ for this biology,” said Patrick Huss, a bioengineer at the Ark Institute and the University of California, Berkeley. He said at a press conference announcing the release of 2.
Other scientists are impressed by how they read about the model. This is described in a paper submitted to the ARC Institute website and submitted to Biorxiv Preprint Server. But they say they need to kick the tire before they can reach a solid conclusion.
“They’re making the most of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the genome of the gen So far, he has been impressed with the engineering behind the model.
Trillions of letters
Over the past few years, researchers trained millions of protein sequences and then predicted protein structures and supported an entirely new design, including the ESM-3 model developed by former META employees. We have developed an increasingly powerful “protein language model.” Proteins containing gene editors and fluorescent molecules.

AI dreams of a new protein blizzard. Do any of them actually work?
Unlike these models, EVO-2 is a genomic data that contains both “coding sequences” with instructions for creating proteins, and a sequence that allows you to control when, where, and how the gene is active. was trained on non-coding DNA containing. The first version of EVO, released last year, was trained on the genome of 80,000 bacteria and archaea (a simple organism called prokaryotes), as well as its viruses and other sequences.
The latest model is based on 128,000 genomes, including the genomes of humans and other animals, plants and other eukaryotes. These genomes contain a total of 9.3 trillion DNA characters. Based on the computing power needed to devour this data and other features, the EVO-2 is the largest biological AI model ever released, says HSU.
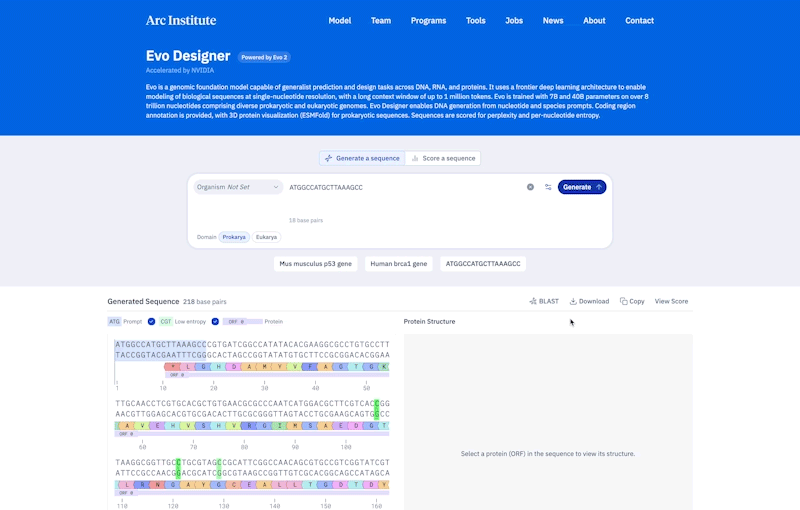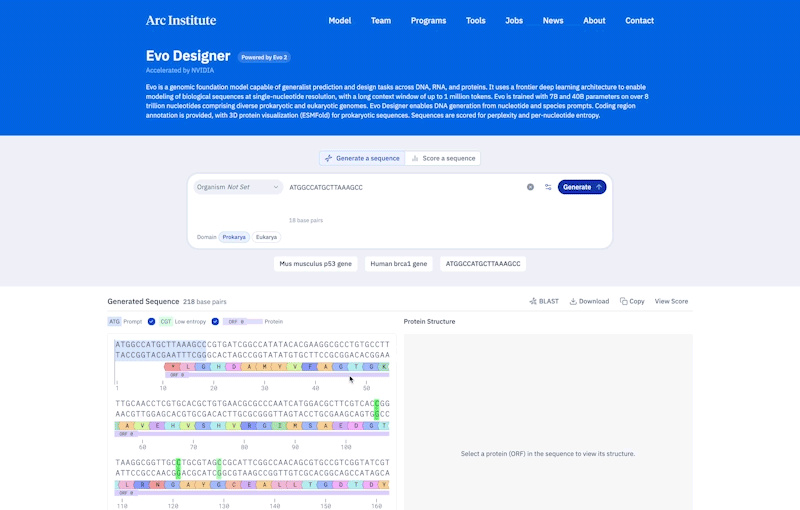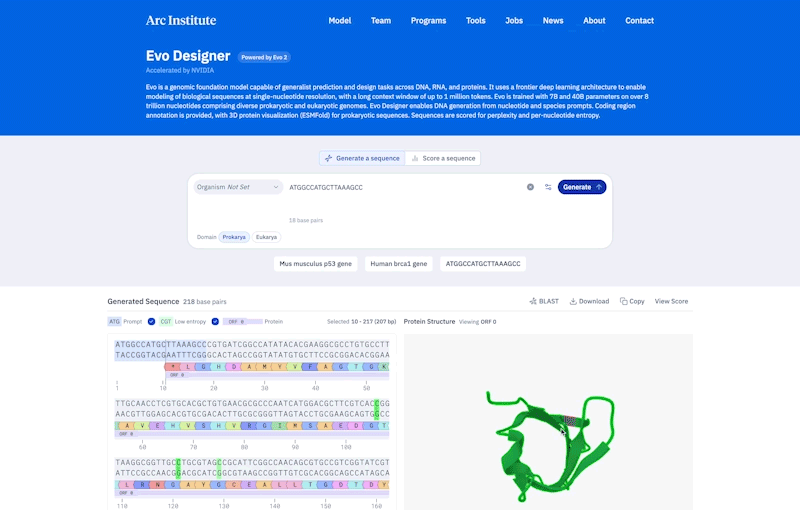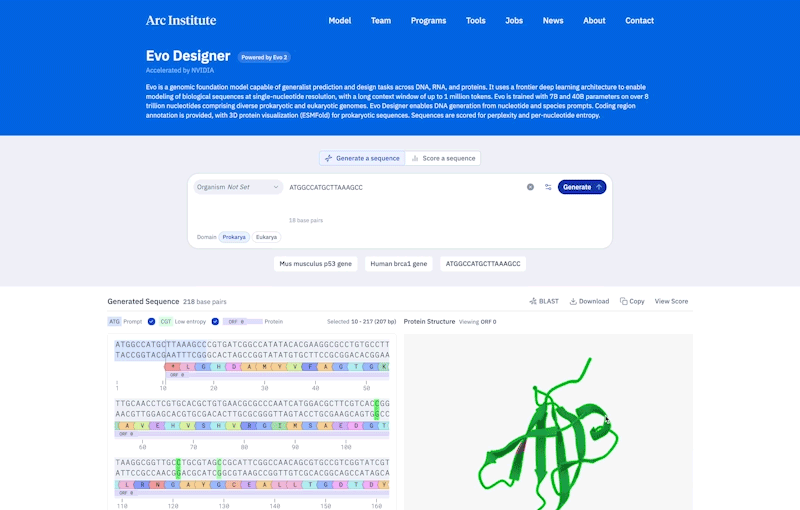
Credit: Ark Research Institute
Compared to prokaryotes, eukaryotes tend to be longer and more complicated. Genes are made of scattered segments of coding and non-coding regions, and the non-coding “regulatory DNA” may be far from the genes they control. To handle this complexity, EVO-2 was constructed to allow learning patterns of up to 1 million pairs of DNA sequences.
To demonstrate the ability to understand complex genomes, HSU and his colleagues used EVO-2 to predict the effect of previously studied mutations in a gene involved in breast cancer called BRCA1. According to HSU, it was done much like a bio-AI model that is best suited to determine whether changes in the coding region cause disease. “It’s the cutting edge of non-coding mutations.” In the future, this model will help identify changes in the patient genome that are difficult to interpret.
The researchers also tested the ability of models to decipher other features of complex genomes, including wool mammoths. “EVO-2 represents an important step in learning DNA-regulated grammar,” says Christina Theodoris, a computational biologist at the Gladstone Institute in San Francisco, California.